Super-resolution microscopy based on projector: High-speed, self polarization modulated three-dimensional digital structured light illumination microscopy imaging
Digital Micromirror Device, known as DMD, is a digital light processing device that has been widely used in projectors in our daily life. It can provide high-speed pattern modulation, bringing technological innovation to the display industry.
Among many remarkable super-resolution microscopy technologies, structured illumination microscopy (SIM) stands out as a gentle and highly compatible approach that can provide ultra-high temporal and spatial resolutions, which can be mainly categorized into two types: two-dimensional structured illumination microscopy (2DSIM) and three-dimensional structured illumination microscopy (3DSIM). However, the axial resolution in 2DSIM is the same as that of wide-field microscopy, leading to significant reconstruction errors when imaging thick specimens with defocus background. In stark contrast, 3DSIM with axial-modulated illumination can effectively double both the lateral and the axial resolution.
Various implementations have been developed to realize the structured illumination and polarization modulation of the 3DSIM system, whose crucial device includes phase grating and ferroelectric liquid crystal spatial light modulator (FLC-SLM). Phase grating with a fixed linear polarizer is rotating and laterally translated to control the direction, phase, and polarization of the structured light, but the imaging speed is limited by the mechanical rotation (~1 s) and transversal movement (~10 ms) of the phase grating. FLC-SLM applies electric fields to the crystal layer and realizes the modulation of the optical field, but it cannot continuously display patterns and the response time of ferroelectric liquid crystal phase retarders is normally milliseconds, which will extremely drag the imaging speed and cause motion artifacts.
In response to the above issues, Professor Xi Peng's team from the College of Future Technology at Peking University has proposed for the first time a high-speed, self polarization modulated three-dimensional structured illumination microscopy imaging method (DMD-3DSIM) based on digital micro-mirror devices (DMD) and electro-optic modulators (EOM), following the successful implementation of laser interference based DMD-SIM (APL 2020) and the open-source 3DSIM reconstruction software Open-3DSIM (Nature Methods 2023). In the recent issue of Advanced Photonics Nexus, a research team led by Prof. Peng Xi in Peking University has reported a new super-resolution microscopy technique that leverages digital display technology to achieve rapid 3D imaging. The research team has built a high-speed auto-polarization synchronization modulation three-dimensional structured illumination microscopy system, by using the auto-polarization-maintenance capability of DMD, in conjunction with the polarization modulation capability of electro-optic modulators (EOM) for the first time. The DMD-3DSIM system yields a 2-fold enhancement in both lateral (133 nm) and axial (300 nm) resolution compared to wide-field imaging, and can acquire a data set comprising 29 sections of 1024×1024 pixels, with 15 ms exposure time and 6.75 s per volume.
Through extensive experimentation, researchers have successfully imaged a series of subcellular structures, including the nuclear pore complex, microtubules, actin filaments, and mitochondria in animal cells. Moreover, they have extended the application of the 3DSIM system to investigate highly scattering plant cell ultrastructures, examining cell walls in oleander leaves, hollow structures in black algal leaves, and features within the root tips of corn tassels. Notably, in a mouse kidney slice, the actin filaments exhibited a pronounced polarization effect, which was effectively captured and visualized in 3DSIM approach.
In addition, the research team has made all the hardware components and control mechanisms openly available in supplementary notes of the article and Github: https://github.com/Cao-ruijie/DMD-3DSIM-hardware. In their commitment to fostering community engagement and enabling wider access to 3DSIM (and related) hardware systems, given the widespread utilization of DMD technology in digital light processing and its cost-effectiveness as a consumer product, this open-initiative will significantly support the community in devising diverse adaptations and iterations, capitalizing on the foundation established by the open-hardware approach. Together with their recently published Open-3DSIM software on Nature Methods (https://www.nature.com/articles/s41592-023-01958-0), the researchers hope that this open-hardware DMD-3DSIM paper will pave a new avenue for the development of 3DSIM multi-dimensional imaging.
The innovative DMD-based 3DSIM technique offers a rapid, precise, and versatile platform for super-resolution imaging, which can facilitate a range of significant biological discoveries and provide a reliable foundation for the advancement of next-generation 3DSIM. In applications of imaging living cells using 3DSIM, the use of brighter and more photostable dyes can extend imaging duration, advanced denoising algorithms can retrieve more information from images with limited signal-to-noise ratio, and currently, fashionable deep learning models based on neural networks can restore and predict 3DSIM images in noisy data in real-time.
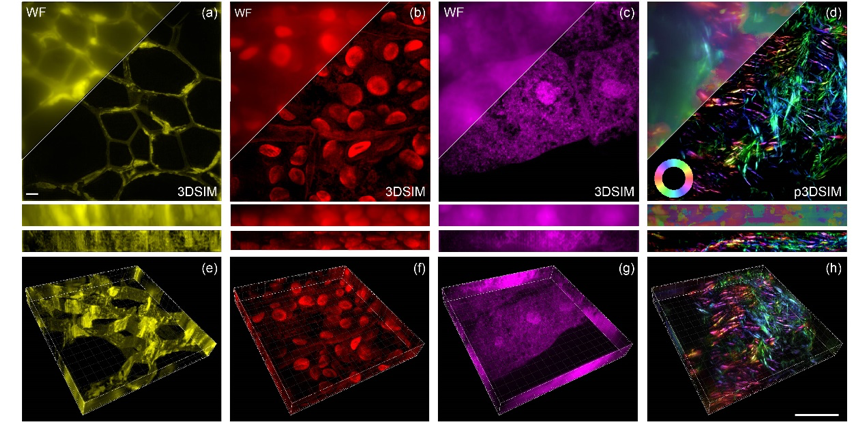
Fig. 1 3DSIM reconstruction of plant and animal tissue samples. The maximum intensity projection (MIP) images of (a) cell walls in oleander leaves, (b) the hollow structures within black algal leaves, (c) the periodic aggregation and dispersion of structures in the root tips of corn tassels, and (d) actin filaments in the mouse kidney tissue slice. The corresponding 3D MIP images are respectively shown in (e-h). 37 layers. Scale bar: 2 μm.
In this work, Ph. D. students Yaning Li and Ruijie Cao from Peking University are the co-first authors, Professor Peng Xi and Dr. Meiqi Li are the co-corresponding authors. This study has been financially supported by National Key Research & Development Program and National Natural Science Foundation of China.
Link: http://dx.doi.org/10.1117/1.APN.3.1.016001




