Nature|Lei Chen's group reports structure of human phagocyte NADPH oxidase in the activated state
Information source: Doctor Lei Chen's team
As early as 1932, it was discovered that leukocytes consume large amounts of oxygen as they phagocytose bacteria[1]. This process is known as the "respiratory burst". In 1957, clinicians identified an inheritable congenital immunodeficiency disease in young children, chronic granulomatous disease (CGD)[2], in which patients suffer from recurrent severe bacterial and fungal infections that result in the development of granulomas at the site of infection. Further studies revealed that neutrophils from these patients, although able to phagocytose pathogens, lacked a respiratory burst and were not able to effectively kill the pathogens they phagocytosed[3]. Subsequent decades of research have shown that the phagocyte respiratory burst is an oxygen reduction process mediated by the NADPH oxidase 2 (NOX2) complex[4]. The phagocytosis of the NADPH oxidase holoenzyme is comprised of two components, a membrane component formed by the membrane proteins NOX2 and p22, and a cytoplasmic component formed by p47, p67, Rac, and p40. In the resting state, the membrane component itself is not active; when stimulated by pathogenic signals, phagocytes will carry out a series of signaling to phosphorylate p47 and other proteins in the cytoplasmic component, lifting its self-inhibitory state, so that p47 binds to the membrane component, and at the same time, recruiting p67, Rac1 and p40 proteins to the membrane component and eventually activating NOX2 protein. The activated NOX2 protein transfers electrons from NADPH in the cytoplasm across the membrane to the oxygen in the phagocytic vesicles, reducing it to generate superoxide anion, leading to rapid oxygen depletion and a respiratory burst[5]. The superoxide anion generated by NOX2 is further catalyzed by enzymes such as SOD and MPO, generating more potent hydrogen peroxide, hypochlorous acid, and other substances, which are used to kill pathogens encapsulated in phagocytic vesicles.
Although phagocyte NADPH oxidase has an extremely important function in natural immunity, the study of its structural mechanism has been lagging behind. Lei Chen's group reported the complete structure of the NOX2-p22 complex in the resting state in eLife in 2022[6]. In the same year, Genentech's group reported the structure of the NOX2-p22 transmembrane region (TMD) in Nature Communications[7]. However, none of these structures contain the cytoplasmic component, which is the component that activates phagocyte NADPH oxidase. The molecular mechanism by which the membrane component of phagocytic NADPH oxidase is activated remains unresolved.
On February 14, 2024, Lei Chen's group at the National Center for Biomedical Imaging Science (NCBI) published an article entitled Structure of human phagocyte NADPH oxidase in the activated state in Nature, which reported the high-resolution cryo-electron microscopy structure of human phagocyte NADPH oxidase in the activated and resting states.
In the early studies of NOX2, effective fragments of cytoplasmic components p47, p67, and Rac1 were often designed as fusion proteins for NOX2 activation in vitro. However, due to the low affinity of the fusion proteins, they were difficult to bind to the NOX2 membrane components efficiently in electron microscopy samples, which led to the difficulty of capturing the electron density of the cytoplasmic components. To solve this problem, the authors, on the one hand, fused GFP and its nanoantibody (GFPnb) to p22 and the cytoplasmic fraction, respectively, to increase the affinity between them, and on the other hand, optimized the length of the linking sequences and their arrangement between p47, p67, and Rac1, and finally chose to co-express the p22-GFP-p67 fusion protein with NOX2, and the formed complex was able to be efficiently activated by lipid-modified GFPnb-p47-Rac1 (a Q61L mutant that stably binds GTP) with high NADPH oxidase activity. The authors finally obtained both the electron density of NOX2 in the activated state and containing a cytoplasmic component (2.79 Å) and the electron density of NOX2 in the resting state without a cytoplasmic component (2.99 Å) by three-dimensional sorting from the same set of cryo-electron microscopy data.
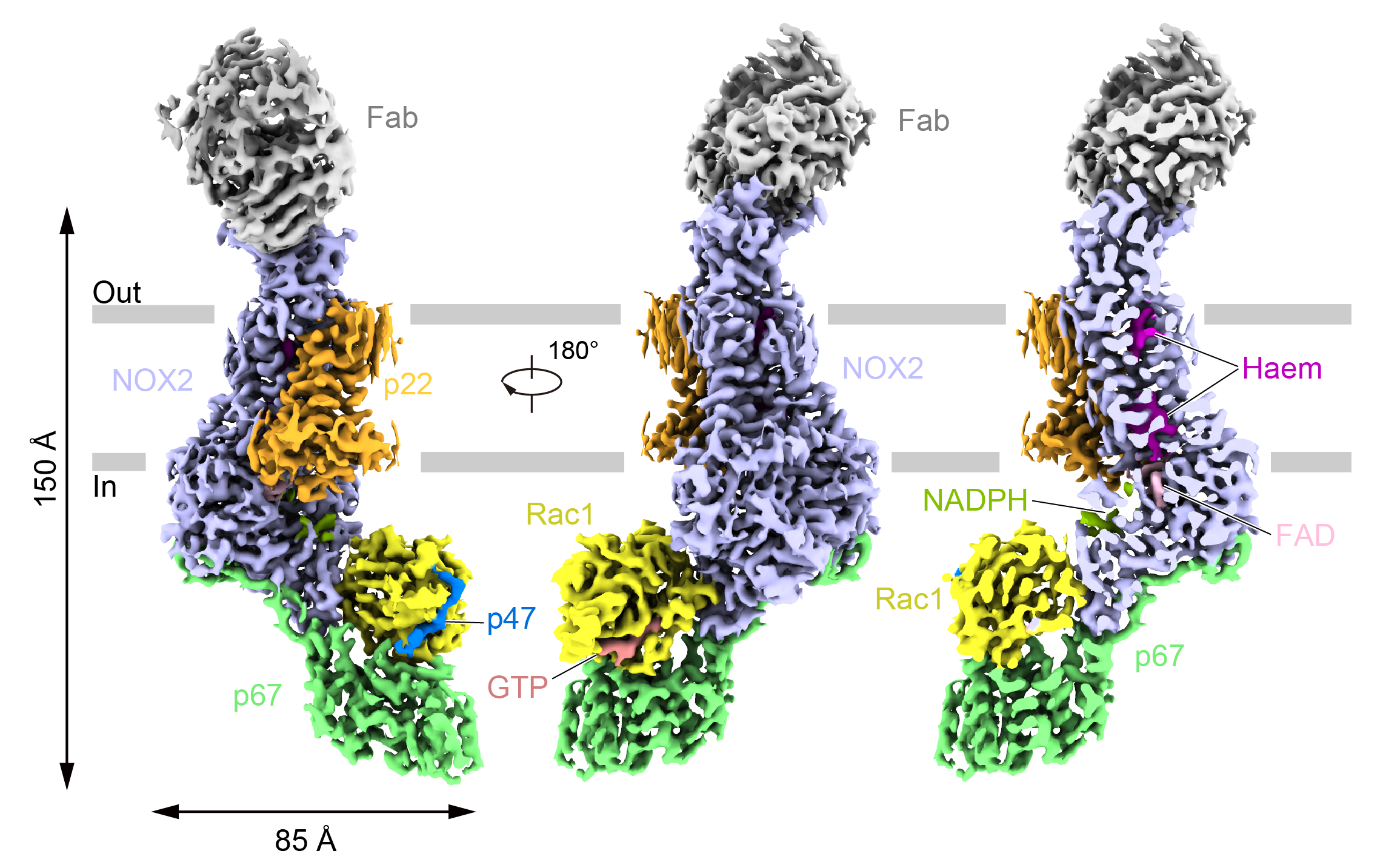
Fig. 1. Electron density map of the NOX2 complex in the activated state
The electron density showed that the activated NOX2 intracellular side was bound to the cytoplasmic components p67, Rac1 and p47, where the transmembrane domains were the classical six-transmembrane helices of the NOX family, and the p67 and Rac1 formed a shape similar to a "mouth", with the p67 representing the "jaw" and the Rac1 representing the "upper jaw", and the two sandwich the dehydrogenase domain (DH) of NOX2 through amino acid interactions. In addition, a small peptide on p47 binds in the cleft between Rac1 α1 and β2.
The binding of p67 to DH relies mainly on the activation structural domain that extends outward from the C-terminus of p67. The β-fold formed by amino acids 202-208 forms an antiparallel arrangement with the β1 of NOX2, and Rac1, which binds to NOX2, is in the GTP-bound state. Mutations of some of the amino acids at the interface of the interaction of DH with p67 and Rac1 reduce the viability of NOX2. Also, several mutation sites found in CGD patients are located near this interaction interface, further suggesting that this interaction interface is critical for NOX2 activation.
By comparing the resting and activated state structures, the authors found that: upon binding of the cytoplasmic component, the DH structural domain of NOX2 docks to the base of the TMD and forms an interaction with the N-terminus of the TMD. The NADPH binds to the DH in the exposed outward cleft, and its adenine and phosphate groups interact with the amino acids surrounding the cleft.
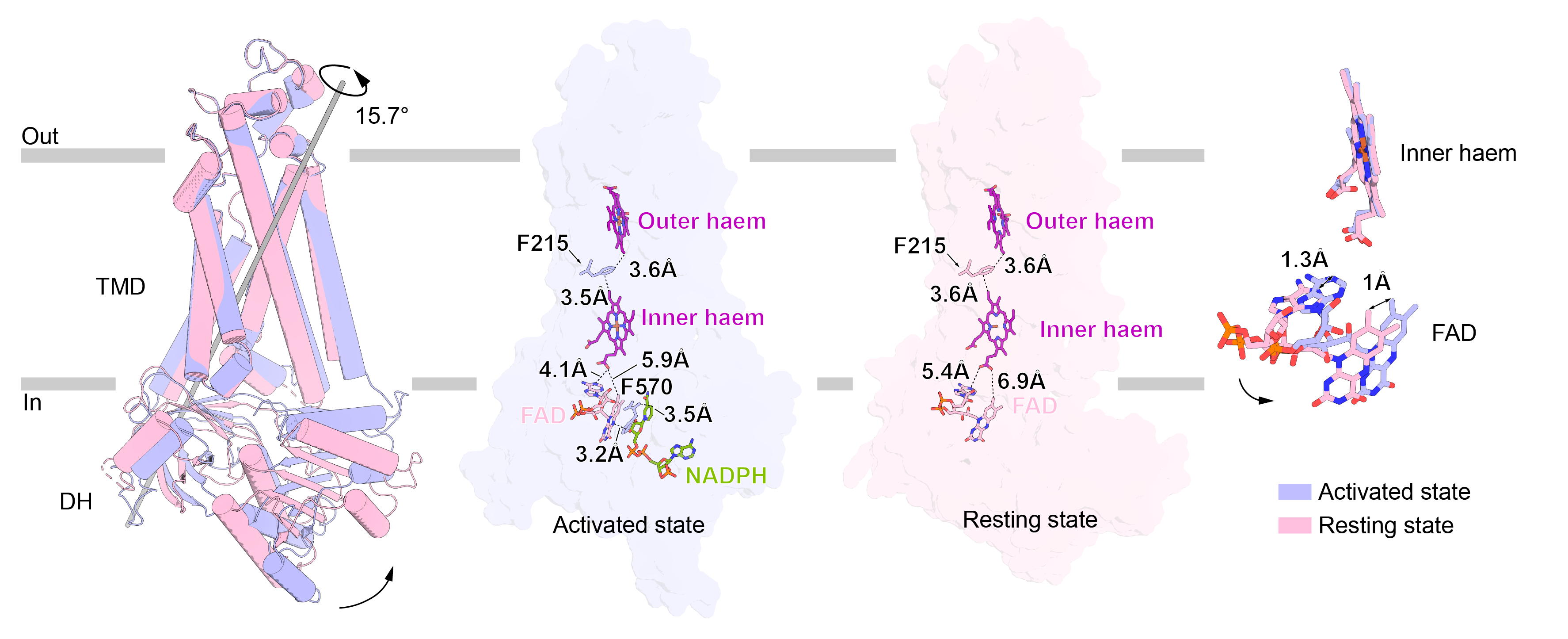
Fig. 2. Conformational changes of NOX2 in activated and resting states
The TMD of NOX2 showed no significant conformational change before or after activation, whereas the DH structural domain was rotated by 15.7° compared to the TMD, forming a more compact conformation and also shortening the distance between FAD and proximal cytoplasmic side of the heme. The FAD-binding structural domain (FBD) produced a 17.4° rotation relative to the NADPH-binding structural domain (NBD), and shortened the distance between the FBD and the NBD distance, bringing R356, which is located on the FBD, closer to NADPH and forming an interaction with it, stabilizing NADPH binding.
By comparing the conformation of NOX2 in both resting and activated states, the authors found that upon assembly of the cytoplasmic components, the binding of p67-Rac1 leads to contraction of the DH structural domain and docking to the TMD, which stabilizes the binding of NADPH and passes its electrons to extracellular oxygen via the FBD, proximo-intercellular hemoglobin, the F215, and proximo-extracellular hemoglobin, ultimately generating superoxide anions.
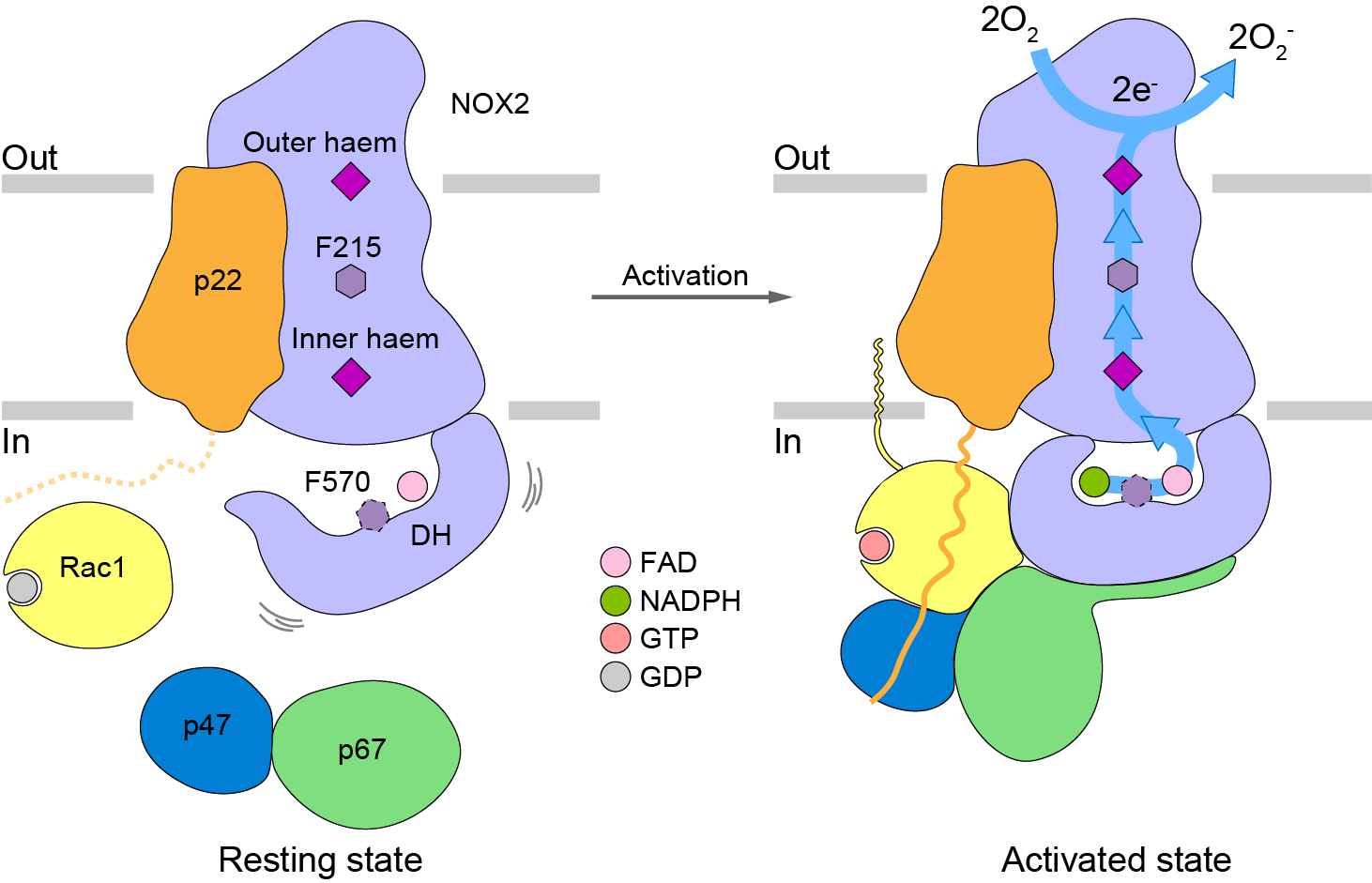
Fig. 3. NOX2 activation model
In summary, the authors used cryo-electron microscopy to analyze the structure of NOX2 in the activated and resting states, and observed at the atomic level how cytoplasmic components p67, Rac1, and p47 bind to NOX2 and cause conformational changes, which provides a structural basis for an in-depth understanding of the activation mechanism of NOX2.
Xiaoyu Liu, a PhD student of the Center for Interdisciplinary Research in Biomedicine (CIRBMB), Institute of Frontier Interdisciplinary Research (IFRIR), Peking University, and Yiting Shi and Rui Liu, PhD students of the School of Futuristic Technology (SFT), were the co-first authors of this work. Dr. Wu Jingxiang (now PI of Concordia Institute of Pharmaceutical Sciences), a postdoctoral fellow, provided important support in the early stage of this work, and Prof. Edgar Pick of Tel Aviv University provided valuable advice on this work. Prof. Jiahuai Han from Xiamen University provided cDNAs of human p47, p67 and Rac1. 7D5 Fab antibody sequencing was done at the Protein Chemistry and Histology Platform of the Protein Research Technology Center, Tsinghua University. The preparation, screening and collection of cryo-electron microscopy samples for this work were done at the Electron Microscopy Laboratory of Peking University, the Cryo-electron Microscopy Platform of the Academy of Biomedical Sciences, and the Songshan Lake Materials Laboratory of the Institute of Physics of the Chinese Academy of Sciences in Beijing, China, with help from Xuemei Li, Zhenxi Guo, Changdong Qin, Xia Pei, Xiao-Juan Hui, Wang Guopeng, and Dapeng Sun. The data processing for this project was supported by hardware and technical support from the CLS computing platform and the Unknown Supercomputing Platform of Peking University.

Figure 4. Group photo of authors (from left to right: Song Kangcheng, Shi Yiting, Chen Lei, Liu Rui, Liu Xiaoyu)
Original link: https://www.nature.com/articles/s41586-024-07056-1s
References:
1. C. W. Baldridge and R. W. Gerard. THE EXTRA RESPIRATION OF PHAGOCYTOSIS. in American Journal of Physiology-Legacy Content, 1932,Vol. 103 235-236.
2. H. Berendes, R. A. Bridges and R. A. Good. A fatal granulomatosus of childhood: the clinical study of a new syndrome. in Minn Med, 1957,Vol. 40 309-12.
3. B. Holmes, A. R. Page and R. A. Good. Studies of the metabolic activity of leukocytes from patients with a genetic abnormality of phagocytic function. in J Clin Invest, 1967,Vol. 46 1422-32.
4. J. D. Lambeth and A. S. Neish. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. in Annual Review of Pathology, 2014,2013/09/21 edn Vol. 9 119-45.
5. C. C. Winterbourn, A. J. Kettle and M. B. Hampton. Reactive Oxygen Species and Neutrophil Function. in Annual Review of Biochemistry, 2016,2016/04/07 edn Vol. 85 765-92.
6. R. Liu, K. Song, J. X. Wu, X. P. Geng, L. Zheng, X. Gao, H. Peng and L. Chen. Structure of human phagocyte NADPH oxidase in the resting state. in Elife, 2022,2022/11/23 edn Vol. 11.
7. S. Noreng, N. Ota, Y. Sun, H. Ho, M. Johnson, C. P. Arthur, K. Schneider, I. Lehoux, C. W. Davies, K. Mortara, K. Wong, D. Seshasayee, M. Masureel, J. Payandeh, T. Yi and J. T. Koerber. Structure of the core human NADPH oxidase NOX2. in Nat Commun, 2022,2022/10/15 edn Vol. 13 6079.




