Peng Xi's Team proposed an open-source 3D structure illumination micro-imaging reconstruction algorithm platform
Source of information: Xi Peng's Team
Due to the advantages of high speed, low phototoxicity and high dye compatibility, structural illumination microimaging (SIM) has become the most powerful super-resolution microscopic technique for dynamic observation of organelles. With the development of SIM, experts and scholars at home and abroad have worked on a series of SIM algorithms, such as open-SIM, fairSIM, Hessian-SIM, HiFi-SIM, etc. These open-source software have also promoted a series of hardware progress, such as SLM-SIM, DMD-SIM, and DMD-SIM. As well as commercial systems such as Airy-SIM, the combination of algorithms and hardware has made the SIM space open and vibrant. In 2008, Gustafsson's research group first proposed the concept of 3DSIM. Compared with 2DSIM, 3DSIM has twice the vertical plane resolution, which greatly eliminates the defocusing background common in 2DSIM and achieves the ability of whole cell imaging. However, due to the complexity of 3DSIM, the development of 3DSIM in general lags behind that of 2DSIM, and its algorithms either exist in closed-source commercial systems or are based on traditional INER-3DSIM, which is less user-friendly and has more artifacts. Therefore, an open source and powerful 3DSIM software urgently needs to be developed to meet the growing development needs of 3DSIM.
Professor Xi Peng is from the Department of Biomedical Engineering, College of Future Technology, Peking University, also from Peking University National Biomedical Imaging Center. After proposing the Open source 2DSIM reconstruction platform Open-SIM (IEEE JSTQE2018), his team developed a new Open source 3DSIM reconstruction platform, Open-3DSIM, published in Nature Methods on July 20, 2023 under the title Open-3DSIM: an open-source three-dimensional structured illumination microscopy reconstruction platform.
In contrast to Open-SIM, Open-3DSIM is built on MATLAB, Fiji and Exe software platforms for users with different needs. It has the advantages of high fidelity and low artifacts, and also has a good reconstruction effect under the condition of low SNR. In addition, the introduction of polarization dipole imaging enables Open-3DSIM to have the capability of polarization imaging.
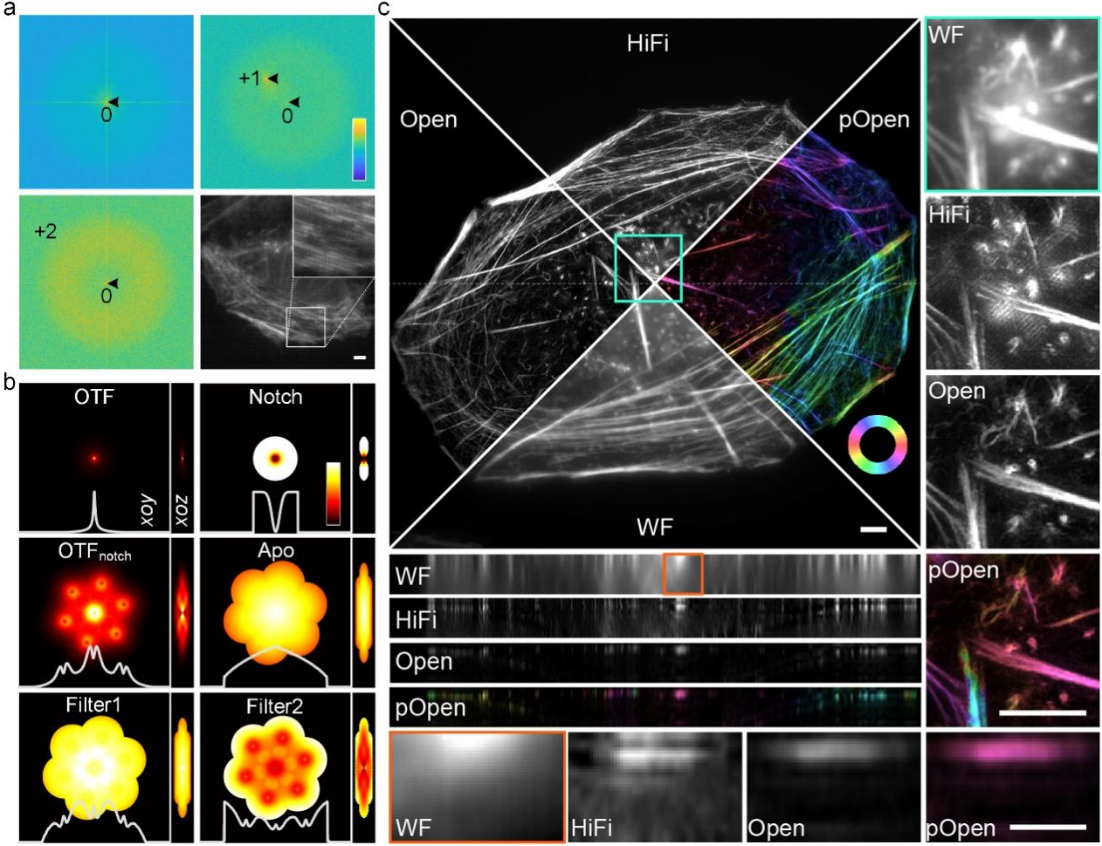
Figure 1. a, collaborative parameter estimation by ±1 and ±2 spectral peaks under low SNR and low-key regime conditions. b, various spectrum optimization filters designed according to the optical conversion function and estimated parameters of the system. c, Comparison between Open-3DSIM and wide-field image (WF), single-layer 3DSIM algorithm (HiFi-SIM) and Open-3DSIM-based dipole orientation analysis (pOpen).
To improve the reconstruction effect of 3DSIM, Xi Peng’s Team proposed an adaptive parameter estimation method, which can greatly improve the accuracy of 3DSIM parameter estimation under the condition of low SNR and low profile. At the same time, a series of frequency domain filtering operations are designed to suppress artifacts and preserve weak details, including notch function to suppress hexagonal artifacts, first-level filtering function to suppress sidelobe and ringing artifacts, and second-level filtering function to preserve details and improve reconstruction resolution. These optimization not only ensure the reconstruction fidelity, but also greatly suppress the defocusing background and various artifacts of 3DSIM reconstruction, making the reconstruction effect of Open-3DSIM comprehensively superior to the existing reconstruction algorithms (AO-3DSIM, 4BSIM, SIMnoise, etc.) and commercial reconstruction software (GE | OMX, etc.). Especially under very low SNR conditions, the mean square error (MAE) and SNR (SNR) of Open-3DSIM and other algorithms are similar under low SNR conditions, which shows the superior reconstruction performance of Open-3DSIM under low SNR conditions.
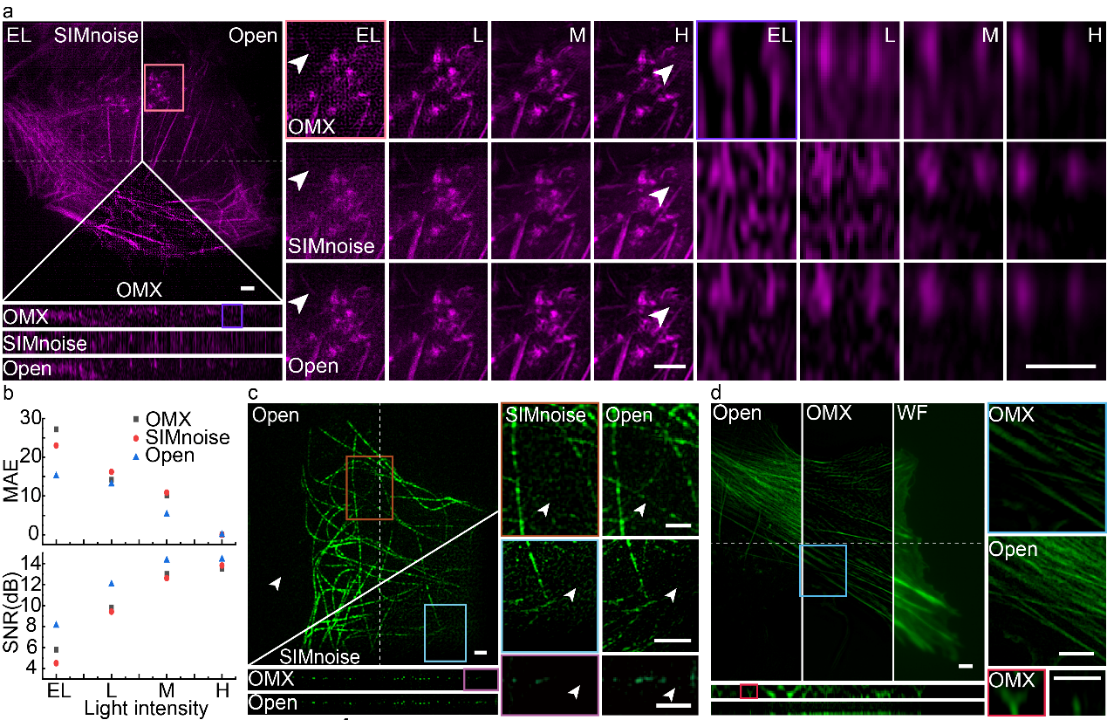
Figure 2. Open-3dsim outperforms existing commercial (GE | OMX) and Open source (SIMnoise) algorithms. A. Analysis of microfilament sample reconstruction results by three algorithms under gradient light intensity excitation (EL, very low light intensity, L, low light intensity, M, medium light intensity, H, high light intensity) and comparison of b, mean square error (MAE) and signal to noise ratio (SNR). c. Comparison of reconstruction of microtubule samples by SIMnoise and Open-3DSIM under extremely low SNR conditions. d, reconstruction comparison of microfilament samples by OMX and Open-3DSIM under extremely low SNR conditions.
According to the principle that fluorescence has different emission intensity of polarized light at different angles, Xi Peng’s team introduced fluorescence polarization orientation information into the post-processing of 3DSIM reconstruction, so that the dipole orientation information of various biological samples can be obtained without any hardware changes. As shown in Figure 3, due to the advantages of Open-3DSIM with low artifacts and high resolution, they successfully performed tricolor imaging of Cos-7 cells, analyzed the three-dimensional structure of mitochondrial ridge and its dynamic separation and fusion process, and obtained the three-dimensional dipole orientation information of microfilament organelles in 5um mouse kidney sections. Open-3DSIM provides transverse/longitudinal superresolution, multi-color, long-term, and dipole-oriented six-dimensional (XYZθT) reconstruction techniques, providing a favorable tool for multidimensional dynamic imaging of suborganelles.
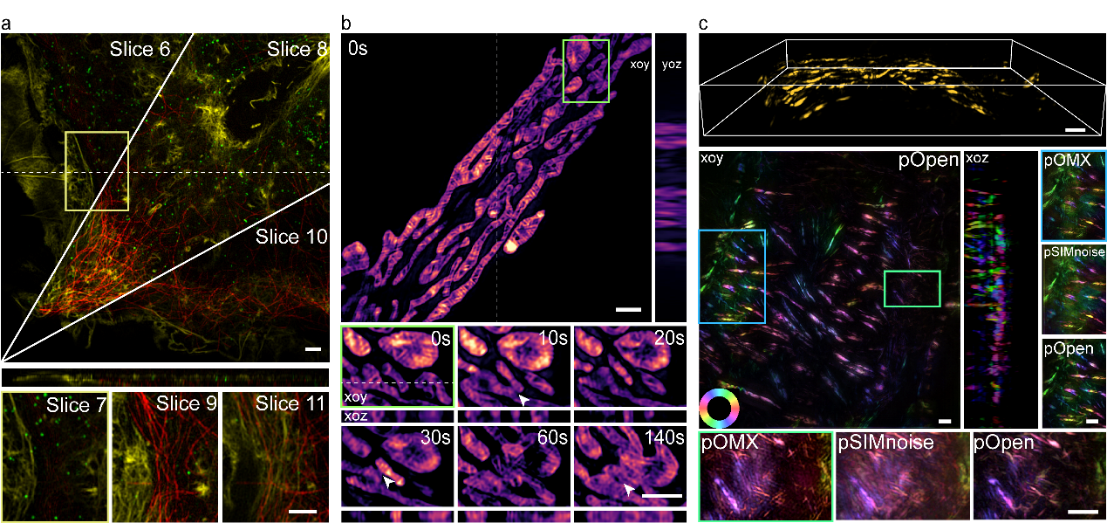
Figure 3. Open-3DSIM establishes horizontal/vertical super-resolution, multi-color, dynamic and polarized six-dimensional imaging modes. (a) Multi-colored Cos-7 cell samples were reconstructed; (b) three-dimensional mitochondrial division, fusion, and apoptosis of Cos-7 cells were analyzed; and (c) fluorescent dipole information of microfilament structure in mouse kidney sections was obtained.
Open-3DSIM is not limited to a single microscope platform, it is widely applicable to GE | OMX, Nikon | N-SIM and other commercial or various homemade 3DSIM systems. It is fully compatible with various existing image optimization methods based on regular/deconvolution/machine learning, and its superior performance, modular design, and extremely user friendliness provide a solid software foundation for the next generation of 3DSIM technology. Prof. Xi Peng hopes that Open-3DSIM will become the international standard for 3DSIM reconstruction to promote the further development of multi-dimensional living cell super-resolution imaging.
Cao Ruijie, a PhD student at Peking University's School of Future Technology, is the first author, with collaborators including Yaning Li, a PhD student at Peking University, Xin Chen, a postdoctoral fellow at Peking University, Xichuan Gai, a master's student at Hebei University, and Meiqi Li, an engineer at Peking University. The authors thank Peking University Life Instrument Platform and Dr. Christophe Leterrier of Aix Marseille University for providing the hardware platform and data for the paper. The authors thank Prof. Hui Li (Suzhou Institute of Medical Engineering, Chinese Academy of Sciences), Marcel Mueller (KU Leuven), and Lin Shao (Yale University) for their careful review and constructive comments. The work was funded by the National Natural Science Foundation of China and the Key Research and Development Program of the Ministry of Science and Technology.
The paper link is: https://doi.org/10.1038/s41592-023-01958-0, disclose the relevant code and reconstruction of the platform in making: https://github.com/Cao-ruijie/Open3DSIM, the authors also open source 3DSIM sample data to Figshare: https://figshare.com/articles/dataset/Open_3DSIM_DATA/21731315, for the use of user testing.




