Cheng Heping's team revealed the system-level time computation and representation in the suprachiasmatic nucleus
Birds flying, butterflies fluttering, hearts beating — these rhythms of life orchestrate the most wondrous symphony of nature. Yet, how do mammals compute time? How precise can such timing be? How many neurons are required to make a robust time signal? Given that a small subset of SCN neurons is critical for maintaining rhythmic function, do SCN neurons contribute uniformly, or else some subpopulations dominate in this process? These questions beg for answers from the scientific community. Recently, an interdisciplinary research team led by Dr. Cheng Heping from Peking University published a research article in Cell Research, which investigated the system-level time-keeping mechanism of the mammalian central circadian pacemaker, the Suprachiasmatic nucleus (SCN). This study reveals that population-level Ca2+ signals predict hourly time via a group decision-making mechanism with modular time feature representation in the SCN.
The team self-built a dual objective two-photon microscope for volumetric Ca2+ imaging of up to 9,000 GABAergic neurons in adult SCN slices. Ca2+ bursts constituted the elemental units to build higher-order temporal features, including “states”, “modes” and “phase wave of hyperactivity” over a circadian period. This demonstrated the potential time-coding capability of SCN Ca2+ signals.
So, how exactly does SCN encode and decode time information? Leveraging machine learning techniques, the team developed a time decoder based on SCN neuronal Ca2+ signals. The decoding accuracy improved significantly with the number of randomly selected neurons, reaching 99.0% accuracy at a cohort size of 900 neurons. This finding suggests a group decision-making mechanism in a statistical system.
Through attribution analysis, the team estimated the contribution coefficients of individual neurons, and found that all neurons have a nearly equalized contribution on a 24-hour scale in terms of time computation.
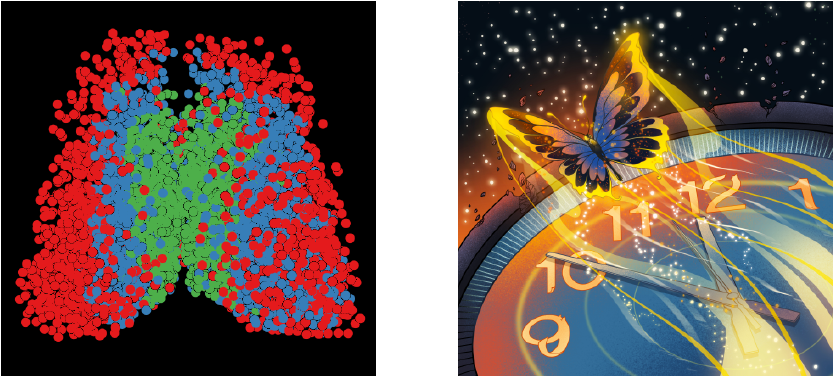
Functional neuron subtypes are organized to form bilaterally symmetrical ripple-like patterns, aesthetically pleasing like that of a butterfly
Further studies revealed that the functional neuron subtypes in the SCN were identified based on a multiscale time series classifier, and elucidated that same-type neurons aggregated in the SCN space, giving rise to distinctive ripple-like patterns with bilateral symmetry, aesthetically pleasing like that of a “Time Butterfly” (illustrated as the above figure).. The modular time representation patterns revealed the nature of invariant properties captured in contrastive learning. The decoding accuracy of the module-specifically learned time decoder can still reach 99%, indicating that different functional modules represent distinctive features.
In summary, the research introduced machine learning techniques to decode signals from large-scale neuronal ensembles, quantitatively characterizing the relationship between the number of neurons polled and time decoding accuracy, and further revealing the time computation capabilities and mechanisms of SCN’s timekeeping at the system level. These findings open a new paradigm in deciphering the design principle of complex neural systems at work based on big data.
Wang Zichen, a PhD candidate from College of Future Technology Peking University, and Dr. Yu Jing (now an associate researcher at PKU- Nanjing Institute of Translational Medicine) are the co-first authors. Dr. Cheng Heping (College of Future Technology), Dr. Hu Wei (Wang Xuan Institute of Computer Science), and Dr. Ma Lei (College of Future Technology) jointly supervised this work. This research was supported by the National Key R&D Program and the National Natural Science Foundation of China, with computational support from Biomedical Computing Platform of National Biomedical Imaging Center, Peking University.
Original link: https://www.nature.com/articles/s41422-024-00956-x




