Dr. Antony K. Chen's group deciphered the roles of RNA scaffolding in the assembly of HIV-1 particles and virus-like particles
Recently, Dr. Antony K. Chen, an Associate professor of Biomedical Engineering at the College of Future Technology, and his group utilized a super-resolution microscopy technique termed photoactivated localization microscopy (PALM) to decipher the mechanisms by which HIV-1 particles and virus-like particles (VLPs) form at the cellular plasma membrane. The research was published in Science Advances with the title: “Roles of RNA scaffolding in nanoscale Gag multimerization and selective protein sorting at HIV membranes” (https://www.science.org/doi/10.1126/sciadv.adk8297).
Gag is the major structure protein of HIV and other retroviruses that can multimerize upon and package a single dimer of the viral genomic RNA (gRNA) or multiple random cellular RNAs, resulting in the formation of a virus particle or a virus-like particle (VLP), respectively. While it is known that both types of RNA molecules serve as scaffolds to drive Gag multimerization, leading to particle assembly, the underlying mechanisms have remained incompletely understood. Using PALM to visualize Gag nanoscale organizations and dynamics in cells with lateral resolution approaching 20 nm, Dr. Chen and his group reported the first evidence of distinguishable gRNA versus cellular RNA-mediated Gag multimerization at the nanoscale (Figure 1). Moreover, these differences in Gag multimerization can have functional consequences for the downstream recruitment of additional plasma membrane (PM) proteins into HIV membranes (Figure 1). Specific major and novel findings are:
1) Compared with gRNA, cellular RNAs are packaged in a less cluster-centric and less cooperative manner, with more sporadic appearances of Gag intermediates exhibiting uncoordinated and perhaps conflicting movements. These differences reflect the ability of the dimer-forming gRNA to act as a long-stranded, continuous scaffold not easily attainable by the multiple random cellular RNAs;
2) While gRNA and cellular RNA-mediated Gag multimerization could result in similar degrees of HIV membrane lipid partitioning, the transmembrane protein murine leukemia virus envelope glycoprotein (MLV-Env), which exhibits a preference to partition into HIV membranes due to its palmitoylated cytoplasmic tail, is two-fold less enriched in the HIV membrane of the gRNA-associated assembly environment compared with the cellular RNA-mediated assembly environment. These results have led us to propose the idea that the high-density gRNA-associated assembly environment imposes greater steric hindrance for proteins with cytoplasmic tails compared to the less densely packed cellular RNA-associated assembly environment.
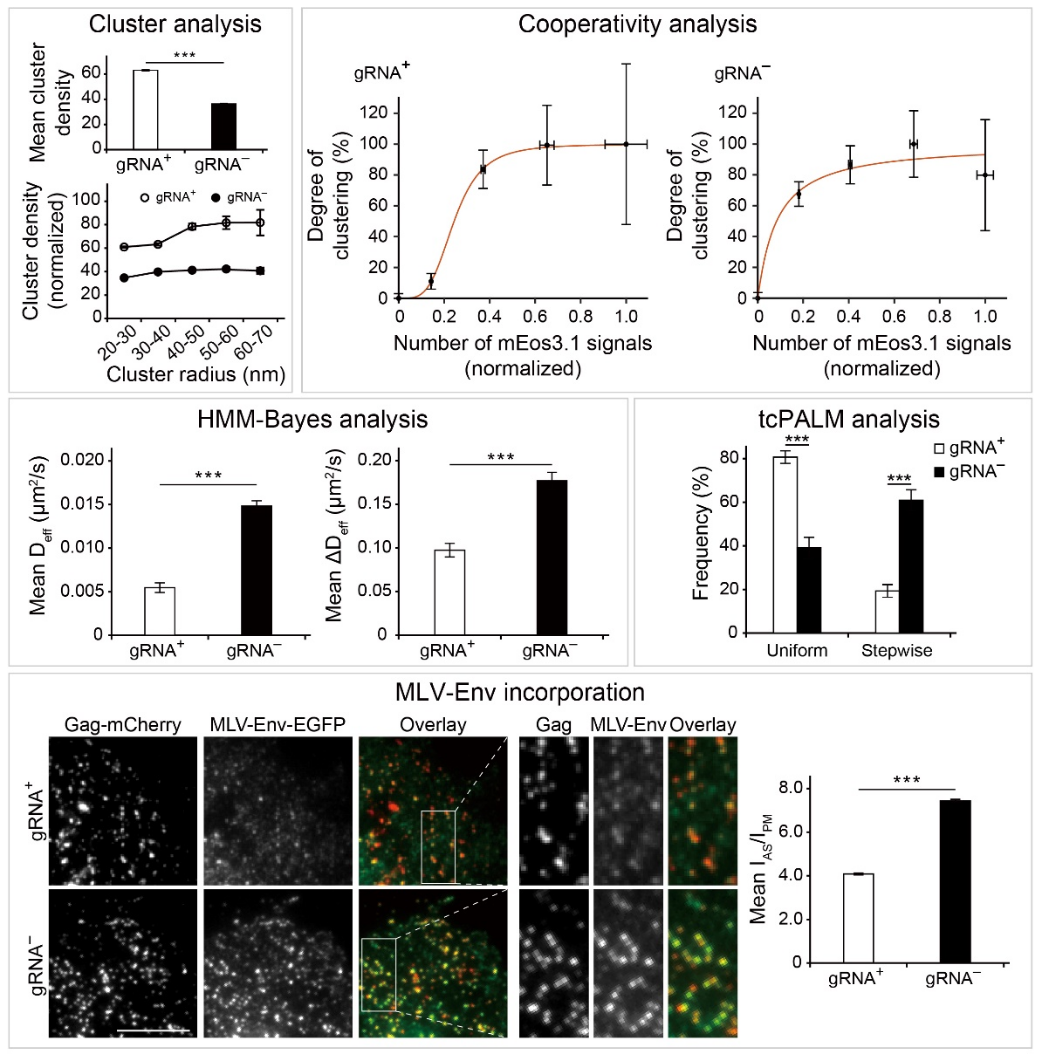
Figure 1. Comparison of Gag nanoscale distributions and dynamics in the presence and absence of gRNA (so that cellular mRNAs are instead packaged). A) Results from PALM. B) More MLV-Env molecules can be incorporated into HIV assembly sites when cellular mRNAs are packaged.
Based on the findings, Dr. Chen and his group proposed the mechanisms by which gRNA versus cellular RNAs mediate Gag multimerization and orchestrate the downstream PM protein sorting into HIV membranes (Figure 2). Specifically, gRNA, which is presumably dimeric following the initiation of Gag multimerization, may serve as a long-stranded modular scaffold to promote Gag multimerization by allowing Gag molecules to interact not only with one another on the same gRNA monomer, but also with those bound to the other gRNA monomer via a cooperative process. The multiple packaged cellular RNAs, on the other hand, function as separate scaffolds that can each only support the multimerization of a subset of Gag molecules, with the cellular RNA-Gag complexes so formed not capable of multimerizing further into larger complexes as efficiently and robustly as when all Gag molecules are connected via a single long-stranded gRNA scaffold. Consequently, the transmembrane protein MLV-Env’s cytoplasmic tail may be more prone to steric interactions with the highly cooperative and densely-packed gRNA-mediated multimerizing Gag complex at the inner PM leaflet, hindering effective recruitment of MLV-Env into the assembly site. It was envisioned that similar mechanisms might also play a role in the recruitment of other transmembrane proteins, such as the HIV-1 envelope glycoprotein, which contains a long cytoplasmic tail that has been reported to interact specifically with Gag.
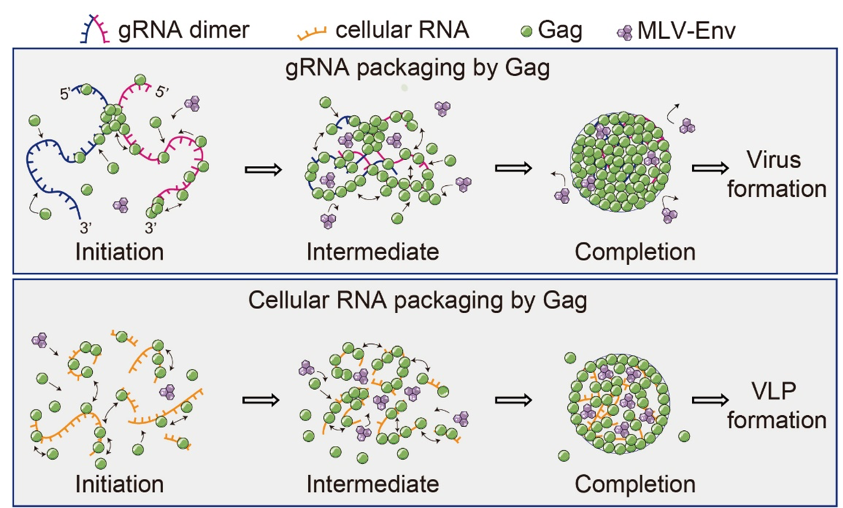
Figure 2. Schematic model illustrating gRNA- and cellular RNA–mediated Gag multimerization and MLV-Env partitioning at assembly sites.
Dr. Chen and his group also believed that the findings that Gag multimerization in the absence of gRNA can result in increased MLV-Env-EGFP recruitment should have important implications in vaccine development involving the engineering of HIV Gag-based VLPs functionalized with viral or nonviral transmembrane proteins as antigens. Specifically, they suggest that VLP-based vaccines might potentially offer greater therapeutic potency compared to conventional live-attenuated and inactivated vaccines due to the ability to accommodate a greater number of antigens owing to the absence of the viral genome.
According to Dr. Chen, “Developments of novel VLP-based RNA delivery and vaccine platforms by my group are currently underway.”




