Sun Yujie's Team analyzes the molecular mechanism of Cohesin complex in 3D genome construction with super-resolution imaging
Information source: Professor Sun Yujie's team
The interphase nucleogenome of mammalian cells presents a highly ordered hierarchical structure. 2m long DNA is continuously compressed and folded, stored in the nucleus, and successively forms different scale domains: Nucleosomes, chromatin fibers, chromatin rings, topologically associated domains (TADs), compartments, and chromosome territory, Each of these hierarchies contains a rich network of DNA interactions that together regulate the cellular life course. The formation and maintenance of genomic hierarchy is regulated by a variety of protein factors, such as architecture proteins, transcription factors and non-coding RNA. The formation of part of the chromatin ring depends on CTCF, cohesin and the loading complex NIPBL-MAU2 and other structure proteins. cohesin makes the chromatin ring grow gradually through "Loopextrusion" and finally forms a stable local interaction domain. CTCF and cohesin combine at its base. However, the specific mechanism by which cohesin and NIPBL work together to promote chromatin extrusion remains unclear.
June 28, 2023, National Biomedical Imaging Science Center (NBIC), Biomedical Frontier Innovation Center (BIOPIC), Peking University College of Future Technology, the research group of Yujie Sun from the State Key Laboratory of Membrane Biology published the title "Rad21 is the core subunit of the cohesin complex involved in directing genome organization's research paper (Figure 1), in which the authors used ultra-high-resolution microscopy imaging techniques to reveal the unique role of the RAD21 subunit of the cohesin complex in cohesin loading and chromatin structure regulation.

Figure 1. Screenshot of the paper
In this study, the team first observed that overexpression of RAD21 causes the chromatin ring to be over-squeezed out, forming a "spaghetti" -like distribution in the nucleus, while the RAD21 protein itself gathers into integrated clusters (Figure 2). Further research shows that the over-loaded cohesin protein forms a "bow-like" structure and binds the TAD (Figure 3), eventually forming a "rosary" distribution, while the overexpression of the other four cohesin subunits is evenly distributed in the nucleus.
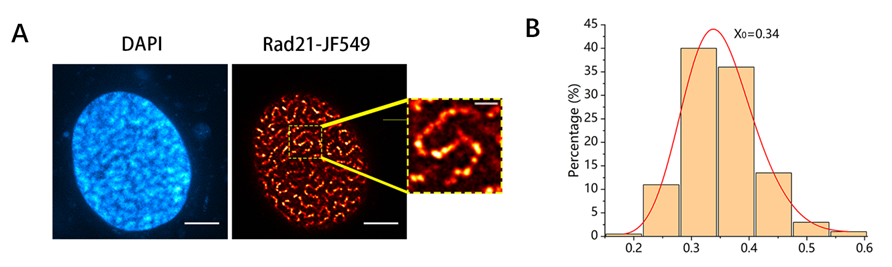
Figure 2. "Spaghetti"-like structure formed in the nucleus after overexpression of RAD21 subunits in HeLa cells
Using the interaction mutant RAD21-LIS of RAD21 and the loading complex NIPBL-MAU2, biochemical experiments and cell imaging experiments confirm the importance of this interaction, which promotes the bulk loading of cohesin on chromatin and finally forms a "spaghetti" -like distribution.
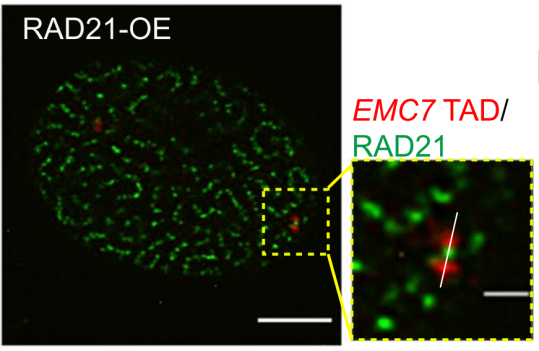
Figure 3. Ultra-high-resolution microscopic imaging of RAD21 and EMC7TAD after RAD21 overexpression
In addition, by analyzing Hi-C data, we compared the effects of RAD21 upregulation on different levels of chromatin (genome-wide, chromatin compartments, and TADs) of WAPL protein deletion, which can also cause "spaghetti" distribution of chromatin, as reported in previous studies. To further explain the mechanism of action of RAD21 on chromatin structure (Figure 4) (Haarhuis et al., 2017; Wutz et al., 2017). The authors found that after the formation of the "spaghetti" -like distribution, a large number of interaction signals were accumulated at the edges and corners of the TAD, and the interaction between TAD was significantly increased. However, the upregulation of RAD21 had no significant effect on the length and number of TADs, nor did the degree of insulation at the boundary between TAds change significantly, which is different from the results caused by the absence of WAPL. The authors speculate that the upregulation of RAD21 may make different TADs more stably separated, so the length, number and insulation coefficient of TADs are not significantly affected. This may be the difference between its mechanism of action and WAPL, which needs to be further studied.
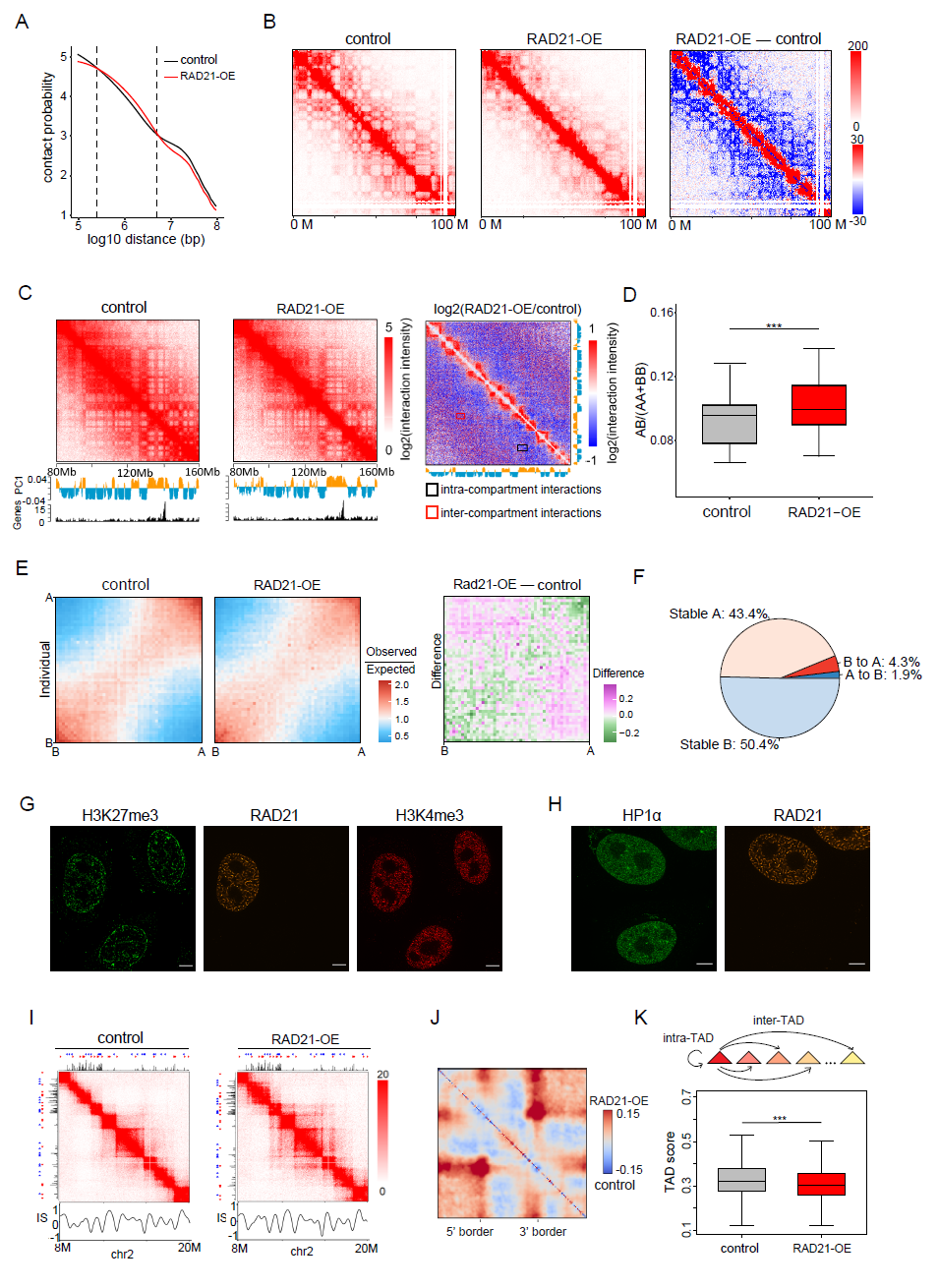
Figure 4 Hi-C data analysis of the effects of RAD21 upregulation on different chromatin hierarchy
Finally, combined with clinical data and cell experiments, the authors found that decreased survival in breast cancer cases was strongly correlated with abnormal expression of RAD21, and immunofluorescence imaging in breast cancer cell lines showed that endogenous RAD21 showed a similar cluster distribution to that in HeLa cells overexpressing RAD21. Combined with RNA-seq and Hi-C analysis, the authors found that upregulation of RAD21 in HeLa cells leads to chromatin compartment switching and significant upregulation of cancer-related genes, and this abnormality of genomic structure may contribute to the development of breast cancer tumors.
To sum up, this study further studies the molecular mechanism of RAD21 subunit promoting cohesin loading process, and proposes a model of how cohesin and loading complex cooperate and promote chromatin extrusion (FIG. 5), which is of great significance for the construction of three-dimensional genome structure in cells. In addition, this study describes the important relationship between chromatin status and cancer etiology, which provides an important reference for clinical treatment target information.
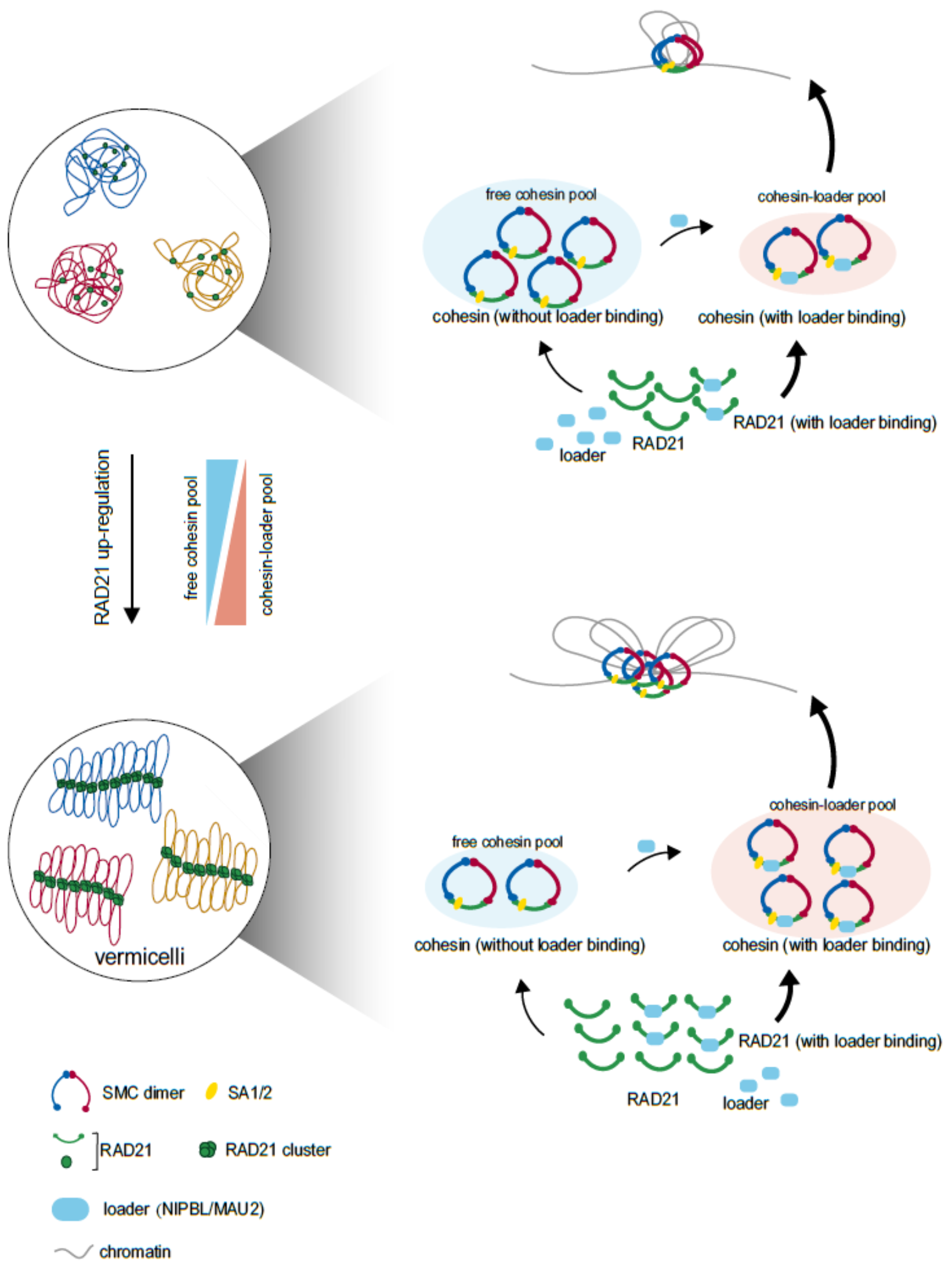
Figure 5. Model of RAD21, as the core subunit of cohesin, which helps the formation of intracellular "spaghetti"-like distribution
Doctoral students Yuao Sun, Xin Xu and Wenxue Zhao from the School of Life Sciences of Peking University are co-first authors of this paper, and Professor Yujie Sun is corresponding author. Laboratory members Chen Keyang, Li Yongzheng, Wang Xiaotian, Zhang Mengling, Xue Boxin and Yu Wanting also made important contributions to the paper. Professor Wang Shu and Dr. Wang Chaobin from Peking University People's Hospital collaborated on breast cancer tissue and cells, and Professor Kong Daochun from Peking University's School of Life Sciences provided important support in the biochemical experiments of this study. Professor Li Cheng from the School of Life Sciences of Peking University and Professor Jie Wei from the School of Life Sciences of Tsinghua University and laboratory member Dr. Zhang Yu provided important assistance in the analysis of sequencing data. This research was supported by the National Key Research and Development Program (No. 2022YFA1303103) and the Outstanding Youth Fund of the National Foundation Committee (No. 21825401), as well as the Instrument Center of the School of Life Sciences of Peking University and the Phoenix Engineering Platform.
Links: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-02982-1
References:
1. Haarhuis, J.H.I., van der Weide, R.H., Blomen, V.A., Yanez-Cuna, J.O., Amendola, M., van Ruiten, M.S., Krijger, P.H.L., Teunissen, H., Medema, R.H., van Steensel, B., et al. (2017). The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693-707.
2. Wutz, G., Varnai, C., Nagasaka, K., Cisneros, D.A., Stocsits, R.R., Tang, W., Schoenfelder, S., Jessberger, G., Muhar, M., Hossain, M.J., et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573-3599.




