Nature Methods | Single-cell drug multi-omics unveils epigenetic mechanisms of drug action and resistance
Source: Aibin He's Research Team
The interaction between small molecule drugs and the genome forms the molecular basis for the treatment of a wide range of diseases, including both traditional drugs and targeted therapies. Despite the extensive clinical validation of treatments that target the genome through small molecules, the specific mechanisms underlying their actions remain unclear, and the mechanisms of drug resistance have yet to be fully elucidated. Constructing a multi-omics map of drug interactions with genome-targeting or chromatin-associated proteins is pivotal for deciphering their roles in cancer therapy and the mechanisms of resistance and relapse. To date, only a few technologies can measure the interaction between small molecules and genome at the bulk level【1-3】, while the analysis of small molecule-genome epigenomic interactions at the single-cell multi-omics level has not yet been achieved. Therefore, there is an urgent need for the development of high-throughput, universal single-cell detection technologies capable of mapping drug-genome interactions.
The Aibin He lab from College of Future Technology and the National Biomedical Imaging Center, Peking University, in collaboration with Dr. Shaokun Shu lab, published an article titled "Single-cell EpiChem jointly measures drug-chromatin binding and multimodal epigenome" in Nature Methods on July 18, 2024. The work develops a high-throughput single-cell drug multi-omics technology, named scEpiChem (Single Cell co-assay of Epigenome and small molecule Chemicals). For the first time, scEpiChem enables the simultaneous capture of drug-target interactions with the genome and multimodal epigenome at the single-cell level, providing novel insights into drug responses, functional heterogeneity, and the molecular mechanisms of drug resistance.

He lab has been dedicated to development of new technologies in single-cell multimodal epigenomics. They have established a series of universal and high-precision single-cell ChIP-seq technologies (scitChIP, CoBATCH) and single-cell multimodal epigenomics technologies (CoTECH, uCoTargetX)【4-7】. The scEpiChem method employs a combinatorial indexing and antibody-PAT T7 complex sequential tagmentation strategies to achieve ultra-high throughput drug multi-omics detection. This approach enables the simultaneous capture of drug-target interactions and multimodal epigenomic maps from millions of single cells (Figure 1).
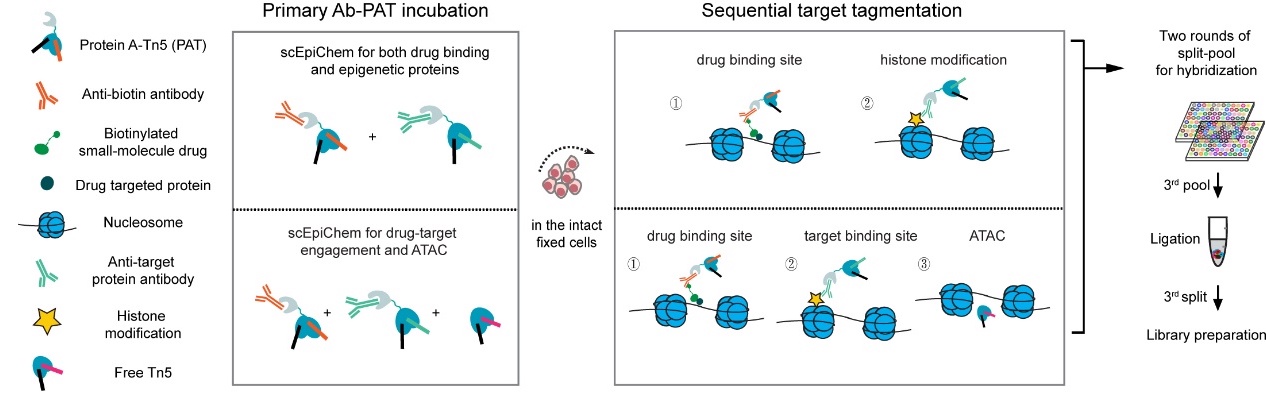
Figure 1. Work flow of scEpiChem
They first employed scEpiChem to simultaneously analyze the BET bromodomain inhibitor JQ1, the CDK7 inhibitor THZ1, and other epigenomic features in cell lines. The results demonstrated that scEpiChem can accurately identify small molecule drug-target binding, histone modifications, and chromatin accessibility across different cell types (Figure 2).
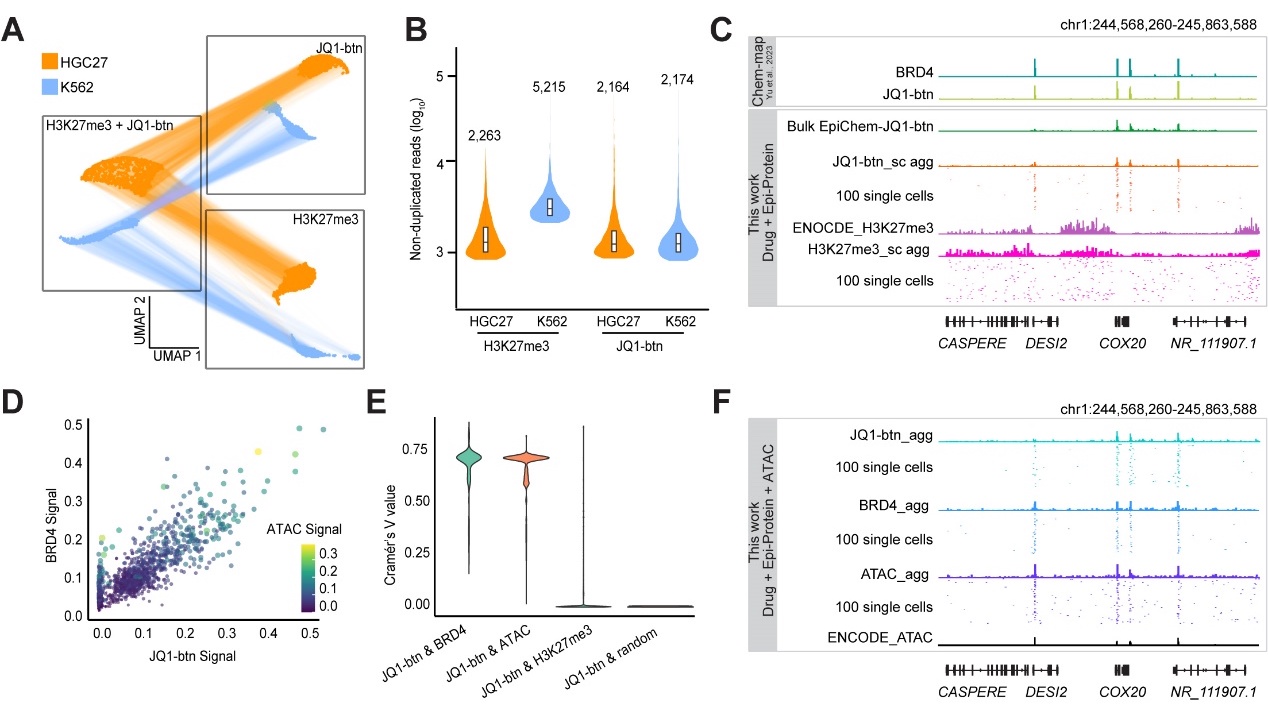
Figure 2. scEpiChem implements a strategy for genome-drug target binding and multimodal epigenetic analysis
scEpiChem provides a comprehensive, high-throughput analytical framework that captures cell heterogeneity in drug responses, which cannot be achieved through traditional bulk analyses. For example, colorectal cancer (CRC) organoids undergoing epithelial-mesenchymal transition (EMT) displayed varying drug treatment responses across different cell types. This approach identified subtle differences between drug-chromatin and target protein-chromatin bindings in each cell type, providing insights into shared and unique binding sites between drugs and their known protein targets (Figure 3).
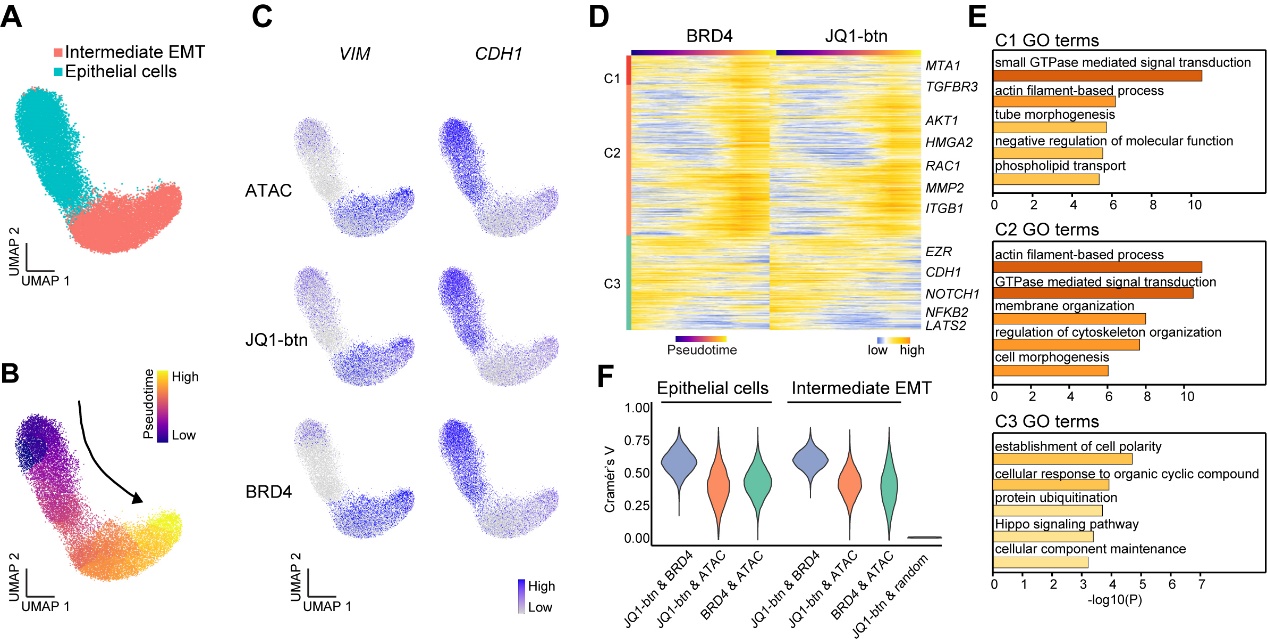
Figure 3, scEpiChem reveals drug response and functional heterogeneity in the tumor microenvironment
PhD Candidates Chao Dong and Tong Zhang from College of Future Technology of Peking University, Xiaoxuan Meng from the Peking-Tsinghua Center for Life Sciences, and postdoctoral fellow Zhifang Guo from Peking University Cancer Hospital are co-first authors of the paper. Drs. Aibin He and Shaokun Shu are co-corresponding authors. Dr. Haiqing Xiong from the Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, made significant contributions to the study. The study was supported by the Ministry of Science and Technology’s Key R&D Program, the National Natural Science Foundation of China, Peking University Chengdu Academy for Advanced Interdisciplinary Biotechnologies, and the Center for Life Sciences.
Original article link: DOI: 10.1038/s41592-024-02360-0
References:
1. Anders, L. et al. Genome-wide localization of small molecules. Nature Biotechnology 32, 92-96 (2014).
2. Tyler, D.S. et al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science 356, 1397-1401 (2017).
3. Yu, Z. et al. Chem-map profiles drug binding to chromatin in cells. Nature Biotechnology (2023).
4. Wang, Q. et al. CoBATCH for High-Throughput Single-Cell Epigenomic Profiling. Mol Cell 76, 206-216 e207 (2019).
5. Ai, S. et al. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol 21, 1164-1172 (2019).
6. Xiong, H. et al. Single-cell joint detection of chromatin occupancy and transcriptome enables higher-dimensional epigenomic reconstructions. Nat Methods 18, 652-660 (2021).
7. Xiong, H. et al. Single-cell joint profiling of multiple epigenetic proteins and gene transcription. Sci Adv 10, eadi3664 (2024).




